Error: Reads File Has Not Allowed Whitespaces in Its First Identifier
Dealing with actress white spaces while reading CSV in Pandas
Why do we care about blank spaces? Build-in pandas functions, and custom handling. Cosmos of 1M examination information and performance test.
![]()
My colleague says that .csv is not an essay so we don't expect whatsoever blank spaces behind the commas (or any other separators). The same stance had the creators of rfc4180 which is commonly understood as a guideline for CSV files. Still, the are inventive developers that feed the .csv exports with unnecessary white spaces. These extra characters are not but increasing the size of our information, simply they can cause much bigger trouble.
In this article we are going to:
- Review why do we care about the spaces in CSVs
- Ready up the benchmark using Pandas's
read_csv()method - Explore the
skipinitialspaceparameter - Try the regex separator
- Abandon the regex separator due to quotes issue
- Apply regex on the loaded dataFrame
- Test the
str.strip()applied column wise on all string columns - Explain why NaN are important
- Generate 1 million lines of examination data using
faker - Measure out the speed and effectiveness in white infinite cleaning of each algorithm
Every bit usual, you can f o llow the lawmaking along with me using the Jupiter notebook containing all the exercises which is downloadable from GitHub.
Why are white spaces a trouble?
- Comparing failures
String with and without blank spaces is not the same. "ABC" != " ABC" these two ABCs are non equal, but the departure is and then pocket-size that you ofttimes don't notice. Without the quotes enclosing the string you hardly would ABC != ABC . But the computer programs are incorruptible in the interpretation and if these values are a merging primal, y'all would receive an empty result.
2. NaN Value
Blank strings, spaces, and tabs are considered as the empty values represented as NaN in Pandas on many occasions. But an indefinite number of spaces is non turned into NaN values automatically and you can get unexpected results, because for example count(NaN) = 0 while count(" ")= 1 .
three. Size
Even though the white spaces are almost invisible, pile millions of them into the file and they will take some infinite. Other fourth dimension they tin can overflow the size limit of your database column leading to an fault in the better example and trimming of the final character whos places was stolen by the bare space in forepart.
How to deal with the white spaces in Pandas?
Let's starting time exploring options we have in Python'southward Pandas library to deal with white spaces in the CSV. As a benchmark permit's simply import the .csv with bare spaces using pd.read_csv() function.
To depict how can we bargain with the white spaces, we will use a 4-row dataset (In club to test the performance of each approach, nosotros will generate a 1000000 records and try to process it at the stop of this article). Our dataset contains these columns:
- Id which identifies each row
- Street which has initial and trailing white space
- Urban center which has leading blank space
- Salary which is numeric
- Date which has a date format
data = """Id,Street,City,Salary,Date
ane, 5th Ave. , Indiana, 100, 2020-07-01
2, Downing Street , San Francisco, 200, 2020-07-02
iii, , New York, 300, 2020-07-03
iv, 1600 Amphitheatre, New York, 400, 2020-07-04
""" You lot can see that the information are containing unnecessary white spaces and our goal is to remove them as finer as possible.
And so that we don't have to shop these data into a .csv file, which we will afterward read, nosotros volition pass it to Pandas using io.StringIO(data).
df = pd.read_csv(io.StringIO(data)) In gild to measure how successful we are, I'll create a function df_statistics() (see below) which iterates through all the columns and calculate:
- Cord columns: total length of data in each row
- Numeric columns: sum of all values
- Other columns: count of rows
Optionally if a dictionary with expected lengths is provided, it volition compare measured lengths or sums with the expectation which I display as a row in a dataFrame. Our benchmark achieves:


def df_statistics(df, expected=None):
"""Calculates the length of all items in the column and decide column types
length of cord
sum of numbers""" res = {}
for col in df.columns:
if pd.api.types.is_string_dtype(df[col]):
# calculate the length of each row and sum it
# careful, you cannot .utilize(len) to columns which contain NAN, because NAN are float. - come across https://stackoverflow.com/questions/51460881/pandas-typeerror-object-of-type-float-has-no-len
fifty = df[col].str.len().sum()
else:
try:# if it doesn't work, it'southward probably numeric value, so allow's only sum
fifty = df[col].sum()
except:
l = df[col].count()
# assigne lenght to the "column_name_lenght" column
res[str(col) + "_lenght"] = l
# if a dict with expected values was provided, allow's employ it to compare with real values
if expected:
res[str(col) + "_diff"] = l-expected[col]
# review the type of the colunmn
res[str(col) + "_type"] = df[col].dtype
return res
Using skipinitialspace
Because .read_csv() is written in C for efficiency, information technology'south the all-time choice in dealing with the white spaces to apply parameters of this method. For this purpose there's skipinitialspace which removes all the white spaces after the delimiter.

Because the Urban center column contained only leading spaces, they were all removed. The concluding row of the Steet column was stock-still as well and the row which contained only two blank spaces turned to NaN, because two spaces were removed and pandas natively represent empty space equally NaN (unless specified otherwise — see beneath.)
But what to do with the blank spaces at the terminate of the rows, betwixt the final character or data and the delimiter. Pandas don't accept whatsoever skiptrailingspaces parameter so we have to use a different approach.
Regular expression delimiters
Did you know that you can use regex delimiters in pandas? read_csv documentation says:
In improver, separators longer than ane character and dissimilar from
'\due south+'will be interpreted as regular expressions and will also force the use of the Python parsing engine. Notation that regex delimiters are prone to ignoring quoted information. Regex case:'\r\t'.
In our case, we can try separator sep="\south*[,]\s*" . \s* mean any number of bare spaces, [,] correspond comma. In Jupiter notebook, you must besides specify engine="pyhton" , because regex separators are processed through python script, not native c-based code. Permit'southward check the results:
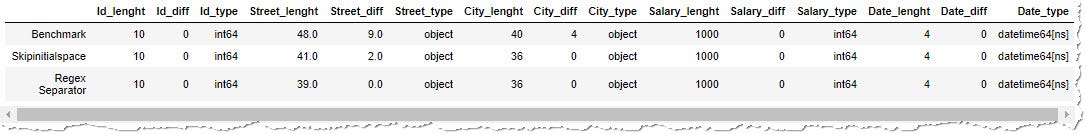
The result seems perfect. All the undesired spaces were removed (all _diff cavalcade equal to 0), and all the columns accept expected datatype and length. But all that glitters is non gold.
Regex separator with quotes
There was an important notation in the manual saying: regex delimiters are decumbent to ignoring quoted information.
Our data were not quoted. Simply if it was, let's say the street would have been quoted:
data = """Id,Street,City,Salary,Date
i," 5th Ave. ", Indiana, 100, 2020-07-01
ii," Downing Street ", San Francisco, 200, 2020-07-02
3," ", New York, 300, 2020-07-03
4," 1600 Amphitheatre", New York, 400, 2020-07-04
""" And then regex separator would not only miss the blank spaces within the quotes, but information technology would also consider the quotes part of the information and our number of extra spaces would even increment:
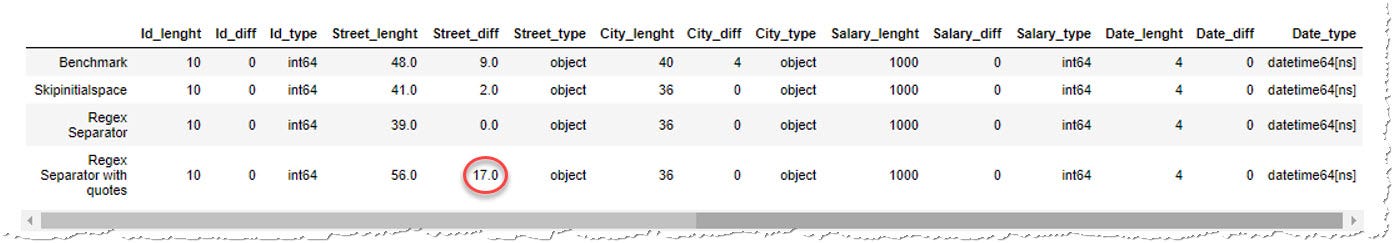
An fifty-fifty worse scenario would happen if the quotes were in that location for a purpose, to shield a separator within the string, in our case comma inside the street name, from existence treated as a separator. " Downing Street, London " protected by the quotes is a valid string containing a comma, but if we use regex separator, it will call up there'south another separator and neglect:
Expected 5 fields in line three, saw 6. Mistake could possibly be due to quotes existence ignored when a multi-char delimiter is used. So the regex separator is actually not an pick.
Applying Regex after
We tin still utilize regular expressions, but simply as a 2d step. Nosotros will carve up the CSV reading into 3 steps:
- read .csv, because the quotes with standard
read_csv() - replace the blank spaces
- after the spaces were removed, transform "" into NaN
In order to hands measure the performance of such an functioning, permit's use a function:
def read_csv_regex(data, date_columns=[]):
df = pd.read_csv(data, quotechar='"', parse_dates=date_columns) # remove forepart and ending blank spaces
df = df.replace({"^\s*|\s*$":""}, regex=Truthful)
# if there remained only empty cord "", alter to Nan
df = df.replace({"":np.nan})
return df
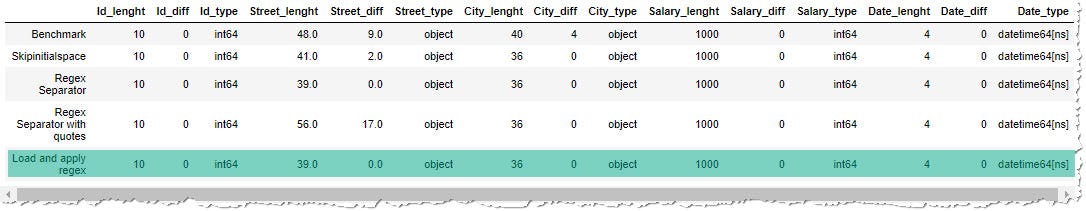
The results are finally encouraging. All the columns have expected lengths and types. But is the operation good?
Strip the strings
Python has a native method to remove the front and end white spaces — .strip() and we can easily utilize information technology on our data. Because it's a string operation nosotros demand to use str.strip() and it can only exist applied on string columns. For that reason, nosotros have to check if the column is having a string format.
# utilize internal method api.types.is_string_dtype to find out if the columns is a string
if pd.api.types.is_string_dtype(df["column_name"]):
df["column_name"] = df["column_name"].str.strip() Over again nosotros wrap the operation into a function so that we tin can apply information technology after in the performance examination. As earlier we will turn all empty string into NaN.
def read_csv_strip(data, date_columns=[]):
df = pd.read_csv(data, quotechar='"', parse_dates=date_columns) # for each column
for col in df.columns:
# check if the columns contains string data
if pd.api.types.is_string_dtype(df[col]):
df[col] = df[col].str.strip()
df = df.replace({"":np.nan}) # if there remained just empty string "", alter to Nan
return df
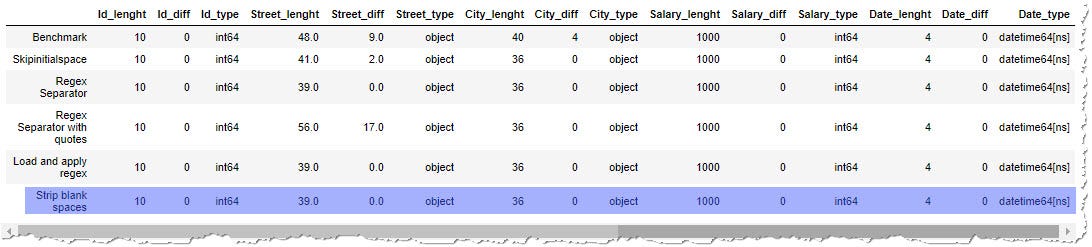
Using str.strip() on the string columns lead to the same quality of the results. Information technology will be interesting to compare the speed of each of the methods.
Note: Don't utilize
df[col].utilise(len)simplydf[col].str.len()becauseapply(len)fails on the NaN values which are technically floats and not strings.
NaN values
Why were we turning all the empty strings into NaNs? By default pandas consider all of these values to be NaNs:
'', '#N/A', '#N/A Northward/A', '#NA', '-one.#IND', '-i.#QNAN', '-NaN', '-nan', 'one.#IND', '1.#QNAN', '', 'N/A', 'NA', 'NULL', 'NaN', 'due north/a', 'nan', 'null'
It has a good reason for information technology because NaN values bear differently than empty strings "".
-
count(NaN) = 0whilecount("")=1 - you can utilize
fillna()ordropna()methods on NaN values
But pandas only turns an empty string "" into NaN, not " " a infinite, two spaces, tab or similar equivalents of the empty space. That'due south why we have to care for whatsoever of these characters separately after the .csv was loaded into the dataFrame.
Performance Test
Generate Exam Data
How fast is each of the suggested approaches? We would presume that build-in methods are the fastest, but they don't fit our purpose. Permit's fix some data in club to meet the real speed of the operations.
A few lines are always candy in a glimpse of an heart, and so we need a meaning corporeality of information in order to examination the operation, let'due south say 1 meg records. For this purpose, I wanted to try python'south faker library which has a quick interface to create random names, addresses, and other data.
import faker
import random # initiate the faker with a seed
f = faker.Faker()
faker.Faker.seed(123) # generate million lines with extra white spaces
data = []
for i in range(one thousand thousand):
data.append([" " + f.address(),
" " + f.name() + " ",
" " + str(random.randint(1,meg))])
I'g creating a list containing a million rows and 3 columns:
- address with actress two blanks spaces at the kickoff
- name with beginning and end white space
- and random number represented as cord with a blank space
The generated dataFrame has the following parameters
[In]: df = pd.DataFrame(information, columns=["Address","Proper noun","Bacon"])
df.info()
[Out]:
<class 'pandas.core.frame.DataFrame'>
RangeIndex: million entries, 0 to 999999
Data columns (total 3 columns):
Address 1000000 non-null object
Name meg non-cypher object
Salary million non-zippo object
dtypes: object(iii)
memory usage: 22.nine+ MB Because the addresses generated by faker contain not only commas just too line suspension, they will exist enclosed in quotes when we export them to the csv. Instance address:
" 1068 Peterson Cape
South Jacquelineville, RI 78829" In that location are many methods which faker offer. If you want a unmarried line address utilise .street_address() for example or .street_address() + ", " + .city() . You can written report the faker documentation here.
Performance on the 1 Milion sample
To be certain, we measured reasonable processing fourth dimension and were not influenced by some peak use of CPU, e.g. when antivirus software runs, we will examination each of the above-described methods 7 times.
import time def process_seven_times(function, params={}):
processing_time = [] for i in range(7):
start = time.fourth dimension()
df = role("test_data.csv", **params)
processing_time.append(fourth dimension.time() - start) return processing_time, df
You can release this block of lawmaking for each scenario, but it's improve to employ the python'south ability to store functions in variables and prepare a dictionary containing all the role and iterate over information technology.
functions = {"No space treatment": {"office": pd.read_csv, "params": {}},
"Skipinitialspace": {"function": pd.read_csv, "params": {"skipinitialspace": True}},
"Regex Separator": {"role": pd.read_csv, "params": {"sep":"\south*[,]\s*", "engine":"python"}},
"Strip blank spaces": {"function": read_csv_strip, "params": {}},
"Load and utilise regex": {"role": read_csv_regex, "params": {}},
"Load and apply regex column wise": {"function": read_csv_regex_col, "params": {}},
} Each fundamental in the dict includes a function and it's parameters — skipinitialspace, separtor, engine etc. The path of the data is always the same so nosotros don't take to repeat it many times as a parameter.
Nosotros will also store the processing times and calculate the statistics using df_statistics(df)
results = []
statistics = [] for name, role in functions.items(): processing_time, df = process_seven_times(function, params=f["params"]) # create a Series with processing time and proper noun information technology based on the role used
s = pd.Serial(processing_time)
s.proper name = name
results.append(southward) statistics.append(pd.DataFrame(df_statistics(df), index=[name]))
Later a few minutes when we test all our functions, we can display the results:
stats_perf_df = pd.DataFrame(results)
stats_perf_df["avg"] = stats_perf_df.mean(axis=1)
stats_perf_df, stats_perf_df["avg"].plot(kind="bar") 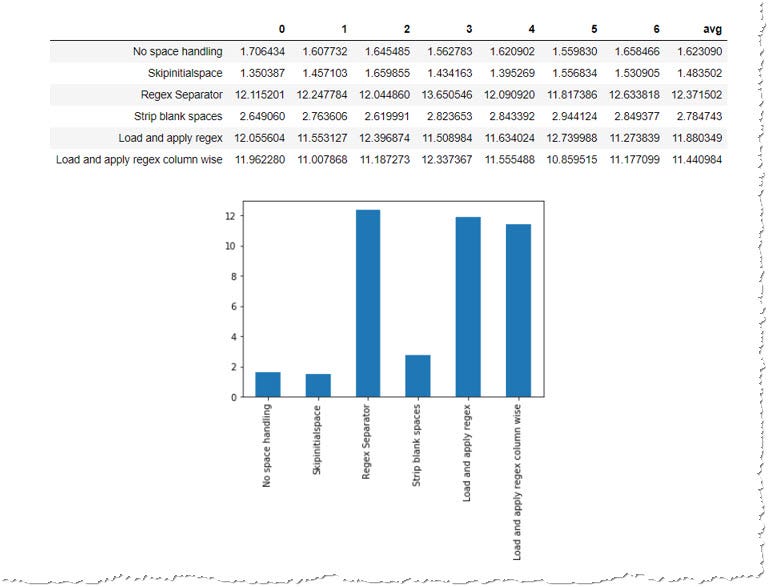
The operation test confirmed what we have expected. Build-in methods trounce custom algorithms. Though since they don't achieve what we want, we tin can employ str.strip() to remove the blank spaces from the loaded dataFrame. It's approx. 50% slower than process without the stripping, just still almost 5-times faster than using the regexes.
You might have noticed that using skipinitialspace can trounce a load without any white space handling, and so combining this parameter with post-processing on the loaded dataFrame tin can bring even better results if speed is our business organization. You lot tin can run the operation examination a second time to confirm that this was not an anomaly.
Conclusion
White infinite treatment is important in case our dataset is polluted with extra spaces non only to decrease the size of the data but mainly to correctly join the data with other sources and to receive expected results of the assemblage of data and NaNs.
Pandas contain some build-in parameters which help with the almost common cases. Any further processing must be done past a custom part which decreases the speed of the procedure. For that reason always try to concur with your data providers to produce .csv file which meat the standards.
You tin perform all the code described in this article using this Jupyter notebook on github. The examination data are non included, but you can generate them through this notebook.
Source: https://towardsdatascience.com/dealing-with-extra-white-spaces-while-reading-csv-in-pandas-67b0c2b71e6a
0 Response to "Error: Reads File Has Not Allowed Whitespaces in Its First Identifier"
إرسال تعليق